[]
sudo mkdir /media/BiotoolsShared-virtual sudo mount -t vboxsf myShare /media/BiotoolsShared-virtual
- remove zero length entries from FASTA
foreach i (*.fsa ) echo $i saco_convert -I Fasta -O tab $i > $i:r.tab sed -r '/\t\t\t/d' $i:r.tab > $i.temp.tab saco_convert -I tab -O Fasta $i.temp.tab > $i rm *tab end
Download genomes[]
- Download genomes from ACC or WGS numbers
for x in CP001859 CP003058 FP929048 HE576794 CP002637 CP001820 ACGB00000000 AFHQ00000000 ACIM00000000 AFBB00000000 AENT00000000 ADGP00000000 AECS00000000 AFUG00000000 AFIJ00000000 ABWK00000000 AEVN00000000 AECV00000000 ACLA00000000 ACKT00000000 AENV00000000 AEEJ00000000 ACKP00000000 AAWL00000000 AEDR00000000 AEDS00000000 ACIK00000000 ADFU00000000 ADCV00000000 ADCW00000000 AENU00000000 AFUJ00000000 do getgbk -a $x > $x.gbk done
- Rename files after organism name in GenBank file and ACC or WGS number
- Procedure copies files
for x in *gbk do extractname $x done
- Remove files named only as ACC or WGS numbers.
- Procedure leaves files with organism names
for x in CP001859 CP003058 FP929048 HE576794 CP002637 CP001820 ACGB00000000 AFHQ00000000 ACIM00000000 AFBB00000000 AENT00000000 ADGP00000000 AECS00000000 AFUG00000000 AFIJ00000000 ABWK00000000 AEVN00000000 AECV00000000 ACLA00000000 ACKT00000000 AENV00000000 AEEJ00000000 ACKP00000000 AAWL00000000 AEDR00000000 AEDS00000000 ACIK00000000 ADFU00000000 ADCV00000000 ADCW00000000 AENU00000000 AFUJ00000000 do rm $x.gbk done
Extract DNA from GenBank[]
- Extract DNA from Origin field of GenBank file
for x in *gbk do saco_convert -I genbank -O fasta $x > $x.fna done
- Rename files from *gbk.fna to *.fna
for x in *gbk.fna do newx=`echo $x | sed "s/.gbk.fna/.fna/"` mv $x $newx done
16S rRNA analysis[]
- Search DNA string for patterns similar to rRNA models in RNAmmer
for x in *fna do echo $x rnammer -S bac -m ssu -f $x.rrna $x done
- Count total number of sequences in a set of rrna files:
grep -c ">" *rrna | awk 'BEGIN{FS=":"}{sum += $2}END { print sum}'
- Rename files from *fna.rrna to *.rrna
for x in *fna.rrna do newx=`echo $x | sed "s/.fna.rrna/.rrna/"` mv $x $newx done
- Extract one 16S rRNA sequence from each genome
- Goes trough each file in the directory called *rrna
select16SrRNA -out all.fna
Selecting the sequence with highest score and length between 1400 and 2000 Default is 1400 - 2000 base pairs, change using -min and -max
Program is writing to file called all.fna Default output file is selected16SrRNA.fna, change using -out
Number of files to be evaluated (extension *.rrna): 32 Number of files with no sequences: 2
Score: 1920.1 :: Length 1545 :: Acidaminococcus_sp_D21_ID_ACGB00000000.rrna Score: 1836.1 :: Length 1557 :: Dialister_invisus_DSM_15470_ID_ACIM00000000.rrna Score: 1878.8 :: Length 1555 :: Dialister_micraerophilus_DSM_19965_ID_AFBB00000000.rrna Unacceptable length: 1325 :: /molecule=16s_rRNA /score=1197.2 :: Dialister_microaerophilus_UPII_345-E_ID_AENT00000000.rrna ............
- Using default setting, 6 sequence do not have the right length (1400-1800)
select16SrRNA -out all.fna | grep Unacc | wc = 6
- Using a smaller minimum length, all 29 genomes have an acceptable 16S rRNA sequence
select16SrRNA -out all.fna -min 1100 | grep -c Unacc = 0
- Performing a multiple alignment of the selected sequences:
clustalw all.fna clustalw all.fna -bootstrap=1000 njplot all.phb
- Save file as post script
Postscript file was made pretty manually in texteditor
Genome atlas[]
- Atlases were created for complete genomes= *For each atlas a folder was created to contain the files for that atlas
- The files are saved but can be deleted as soon as the atlas has been drawn
mkdir GenomeAtlas Vparvula sed s/XXXX/Veillonella_parvula_DSM_2008_ID_CP001820/g /usr/biotools/genomeAtlas > Vparvula.genomeatlas.sh chmod +x Vparvula.genomeatlas.sh ./Vparvula.genomeatlas.sh
Identify open readingframes in DNA using Prodigal[]
for x in *fna do prodigalrunner $x done
Bias in third codon position and Amino acid and codon usage[]
- Calculate AT and amino acid usage in open reading frame files, FASTA format
for i in *orf.fna do perl /usr/biotools/indirect/atStats.pl $i > $i.atStats.tab cat $i.atStats.tab > $i.CodonAaUsage perl /usr/biotools/indirect/CodonAaUsage.pl $i >> $i.CodonAaUsage rm $i.atStats.tab done grep aa *AaUsage > aaUsage.all sed -i s/_prodigal.orf.fna.CodonAaUsage:aa//g aaUsage.all grep Total *AaUsage > statistics.all sed -i 's/_prodigal.orf.fna.CodonAaUsage:/\t/g' statistics.all cut -f2,3,4,5,6,7,8 statistics.all > tmp.all mv tmp.all statistics.all sed -i 's/_prodigal.orf.fna//g' statistics.all grep codon *AaUsage > codonUsage.all sed -i s/_prodigal.orf.fna.CodonAaUsage:codon//g codonUsage.all
Drawing heatmaps of Bias in third codon position and Amino acid and codon usage[]
install.packages("gplots")
library(gplots)
codon <- read.table("codonUsage.all")
colnames(codon) <- c( 'Name', 'codon', 'score', 'count')
codon <- codon[1:3]
test <- reshape(codon, idvar="Name", timevar="codon", direction="wide")
codonMatrix <- data.matrix(test[2:length(test)])
rownames(codonMatrix) <- test$Name
codon_heatmap <- heatmap.2(codonMatrix, scale="none", main="Codon usage", xlab="Codon fraction", ylab="Organism", trace="none", margins=c(8, 25))
dev.print(postscript, "codonUsage.ps", width = 25, height=25)
dev.off()
library(gplots)
aa <- read.table("aaUsage.all")
colnames(aa) <- c( 'Name', 'aa', 'score')
test <- reshape(aa, idvar="Name", timevar="aa", direction="wide")
aaMatrix <- data.matrix(test[2:length(test)])
rownames(aaMatrix) <- test$Name
stat_heatmap <- heatmap.2(aaMatrix, scale="none", main="Amino acid usage", xlab="Amino acid fraction", ylab="Organism", trace="none", margins=c(8, 25), col = cm.colors(256))
dev.print(postscript, "aaUsage.ps", width = 25, height=25)
dev.off()
Bar and rose plots for amino acid, codon usage and bias in third position[]
for x in *orf.fna do basicgenomeanalysis $x /usr/bin/gnuplot done
BLAST martix[]
- Input file is created in directory with protein files in FASTA format
- Takes all fsa files in directory
makebmdest . > bmdest.xml blastmatrix -cpu 5 bmdest.xml > blastmatrix.ps
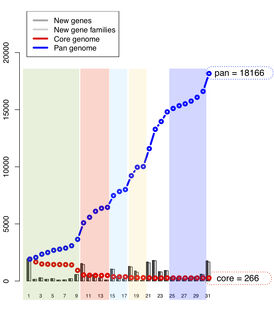
Core and pan genome analysis, single linkage clustering[]
- Input file is created in directory with protein files in FASTA format
- The output is a post script file and a table written to the screen =
ls -1 *orf.fsa | gawk '{print $1 "\t" $1}' > pancore.list
pancoreplot pancore.list
Genome statistics[]
for x in *orf.fna do genomeStatistics $x >> all.stats done